Structure Based Enzyme Engineering (SBEE)
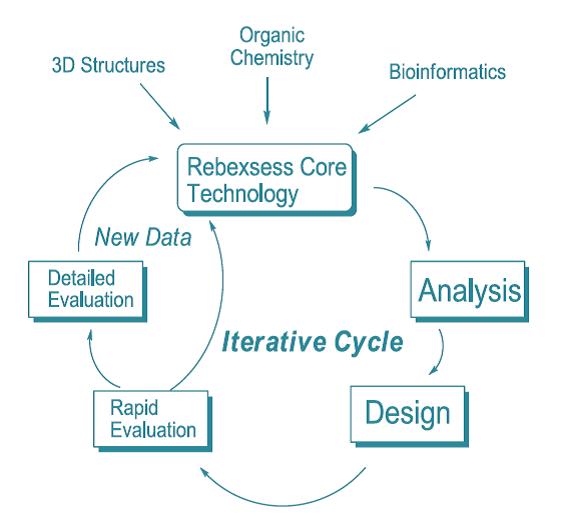
Structure Based Enzyme Engineering is the use of protein structure & enzyme mechanism to create novel enzymes. Rebexsess has unique expertise for the rational design of novel enzymes, not found in nature, useful for catalyzing important reactions. Rebexsess has unique capability to offer 3D protein structure based enzyme design for applications in biocatalysis and enzyme/protein engineering, which integrates computational modeling and organic chemistry into synthetic biology. Rebexsess expertise is especially effective to debottleneck along a pathway or to optimize an individual enzyme in a biosynthetic system.
These transformations can be used to create high value targets such as pharmaceutical intermediates or “green routes” to commodity chemicals. A Rebexsess advantage is the seamless integration of the key disciplines of Organic Chemistry, Protein Structure and Function, and small molecule Enzyme Inhibitors and Molecular Recognition of protein-ligand complexes integrated with practical Computational Chemistry. As in drug discovery, high quality models are a key for advancing rational structure-based programs in enzyme engineering. Rebexsess expertise is used to develop high quality 3D models of enzyme mechanism and putative roles for key protein residues. In SBDD models based on protein-ligand complexes are used to design new and better ligands. In SBEE models based on protein-ligand complexes are used to design better enzymes. Thus, the same key tools and intuition important for using protein structure in drug discovery apply to research in biocatalysis.
Services and Feature of Structure Besed Enzyme Engineering
Modeling Approaches
The Computational Modeling Impact to a Process of Enzyme Engineering: